Aggregation of ubiquitinated cargo by oligomers of the protein p62 is an important preparatory step in cellular autophagy. In this work a mathematical model for the dynamics of these heterogeneous aggregates in the form of a system of ordinary differential equations is derived and analyzed. Three different parameter regimes are identified, where either aggregates are unstable, or their size saturates at a finite value, or their size grows indefinitely as long as free particles are abundant. The boundaries of these regimes as well as the finite size in the second case can be computed explicitly. The growth in the third case (quadratic in time) can also be made explicit by formal asymptotic methods. In the absence of rigorous results the dynamic stability of these structures has been investigated by numerical simulations. A comparison with recent experimental results permits a partial parametrization of the model.
Bioessays 2023 Oct;45(10):e2300044
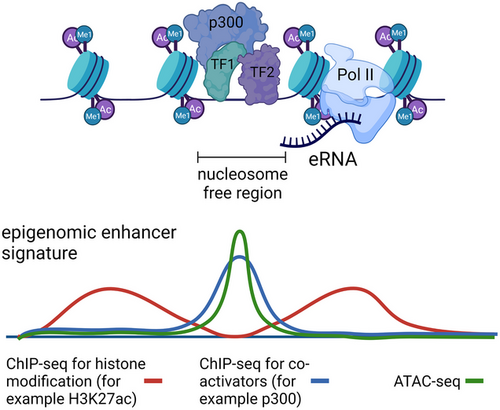
H.F. Thomas, C. Buecker
Bioessays 2023 Oct;45(10):e2300044
H.F. Thomas, C. Buecker