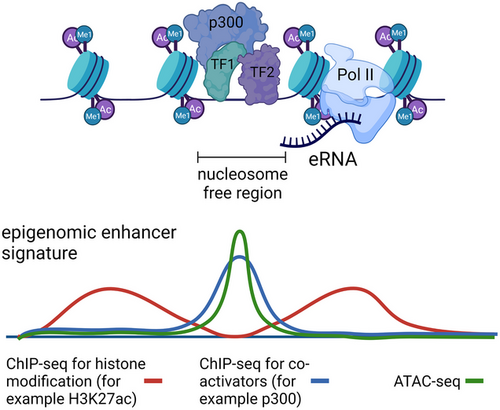
Many genes are regulated by multiple enhancers that often simultaneously activate their target gene. However, how individual enhancers collaborate to activate transcription is not well understood. Here, we dissect the functions and interdependencies of five enhancer elements that together activate Fgf5 expression during exit from naive murine pluripotency. Four intergenic elements form a super-enhancer, and most of the elements contribute to Fgf5 induction at distinct time points. A fifth, poised enhancer located in the first intron contributes to Fgf5 expression at every time point by amplifying overall Fgf5 expression levels. Despite low individual enhancer activity, together these elements strongly induce Fgf5 expression in a super-additive fashion that involves strong accumulation of RNA polymerase II at the intronic enhancer. Finally, we observe a strong anti-correlation between RNA polymerase II levels at enhancers and their distance to the closest promoter, and we identify candidate elements with properties similar to the intronic enhancer.
Bioessays 2023 Oct;45(10):e2300044
H.F. Thomas, C. Buecker
Bioessays 2023 Oct;45(10):e2300044
H.F. Thomas, C. Buecker